PERDIX: an algorithm for the automatic design of DNA origami of different geometries
Who did not like to collect designers in childhood? I still remember this red box with a bunch of metal parts, tools and a sea of possible results if only there was imagination, time and desire. LEGO also should not be forgotten, even though everything was a bit simpler and much more colorful. But what is more complicated is that both are the designers of nanostructures based on DNA origami. Until now, all the parts of such structures were modeled manually, which took a lot of time and effort. Imagine that you need to create all the LEGO parts yourself before you assemble them into a giant robot with lasers, jet engines and a machine gun on your shoulder. But something childhood memories took us to the wrong steppe.
Seriously. Today we will get acquainted with the algorithm that allows you to automatically create DNA origami of a fairly diverse form. Previously, changing the shape of the DNA strands to the necessary was done manually, which greatly limited the possibilities of such a procedure. This same algorithm allows you to create the details of the DNA constructor automatically, which allows you to further use them for the formation of two-dimensional and three-dimensional nanostructures. Plus, this algorithm is available to everyone. A research group report will help us figure out what and how it works. Go.
The basis of the study
The use of DNA as a building material in nanostructures of a wide range of applications in recent years has become the goal of many studies and experiments. Many of them have achieved excellent results, but as we know, there is no limit to perfection. As an example, researchers cite monodisperse DNA structures on the megadalton scale with a high coagulation factor, which was realized by means of long single-stranded DNA ssDNA frameworks. However, in this case, it is the length of the DNA strands that is the main limitation of the size of a single DNA origami. This restriction can be circumvented by the formation of a structure consisting of several origami at once.
Also, we already know ( Tic-tac-toe: demonstration of the controlled process of DNA structure reconfiguration ) that it is possible to create some DNA structures by means of DNA tiles, which allows us to form complex geometry of this structure itself.
But again, everything depends on the fact that the process is performed manually, and this is a long and dreary. What do scientists like? That's right, automation, or at least partial automation. Such tools already exist (for example, scientists - caDNAno or Tiamat), but these programs have their drawbacks. First, it’s still a manual framing. Secondly, the limitations of the above-mentioned algorithms in the matter of systematization of the design of the geometry of the structure.
If in a nutshell, then in this work, the researchers set a goal to create an algorithm that will do almost everything independently. Thus, any person can set only the parameters of the outer shell of the desired structure, and the program will fill the inside independently with all the necessary DNA with threads, staples, sequences and other things. It is also possible to carry out the process in the reverse order - to set all the “insides”, so to speak, the grid of the structure, and the algorithm will construct the correct and most optimal framework around it.
This open source program (PERDIX) is available to absolutely everyone, for which thanks to the researchers. Thus, anyone can not only “play around” with the program itself, but also, perhaps, make some improvements to its code. Where to download it and how to start we will look at the end of the article.
Now let's look at the examples of the application of this program, which the research group has provided us.
PERDIX Test Results
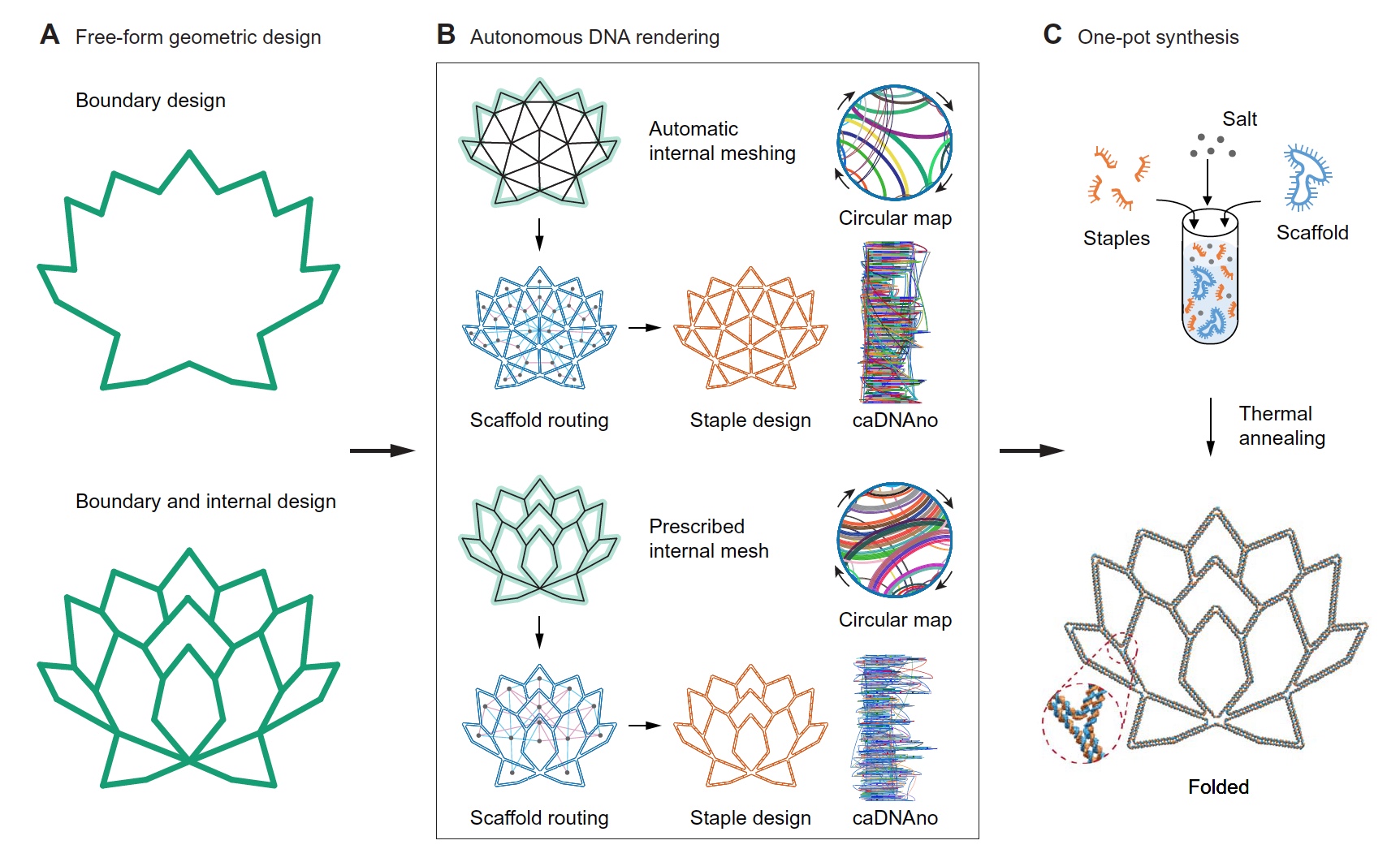
Image number 1
As input data, either an image of the boundary of the desired structure ( 1A , above), or a drawing with more accurate geometric details ( 1A , below) is used. Further, the program itself fills the internal space.
When only an “outline” is specified, the internal geometry of the grid of the structure is formed by the triangles through the open source program DistMesh, which requires the introduction of only the density of the grid as a necessary parameter.
When the shape and geometric lines are specified ( 1A , below), Shapely is used - a Python package for manipulating and analyzing flat geometric objects. Shapely generates a polygonal mesh in which its lines and their points of intersection form the required framework geometry.
Further, after determining the desired shape, it was necessary to specify the nominal edge length, which should be ≥ 38 bp (paired bases) or 12.58 nm. Thus, the required structure will unambiguously contain at least two paired crossings for each edge.
Then all the target lines of the future structure are converted into DX edges, in which all vertices are multi-branch connection points ( 1B ). Following the principles of graph theory, an optimal framework structure is automatically formed, after which the process of assigning a sequence of additional DNA straps is completed. The process of manually editing the output (results) is carried out through the generated caDNAno file.
Scientists call a very important feature of their system the fact that the ribs do not have to be equal to the whole number of B-DNA double helix (10.5 bp). And this allows you to get more freedom in the design of the geometry of the structure.
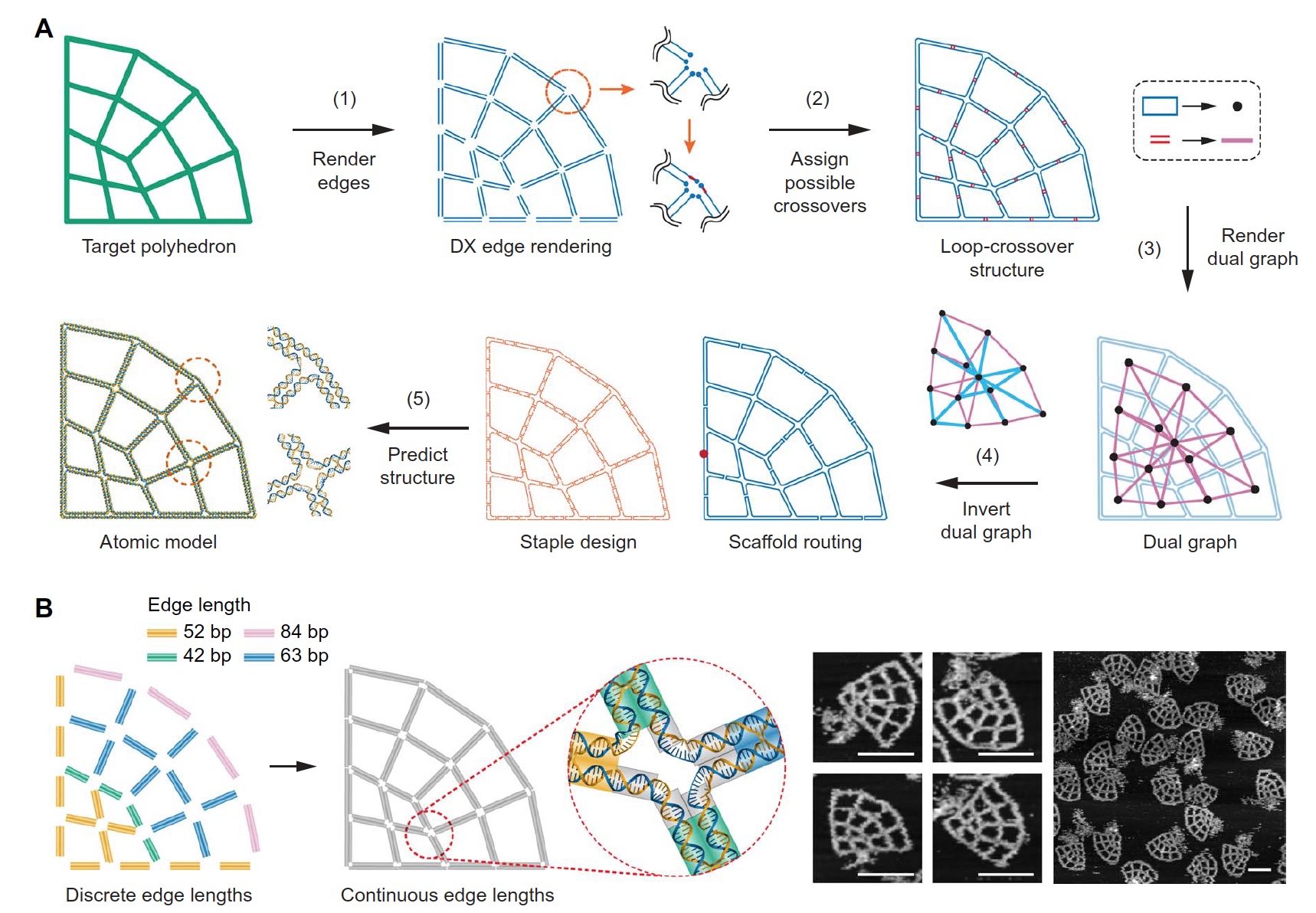
Image number 2
In order to quickly and accurately perform the automatic part of the process of forming the geometry of the structure, all edges must first be converted into DX motifs by rendering each edge of the target geometry using two anti-parallel framework lines ( 2A ). Then they are united at a single vertex, which makes each edge part of a loop. And this in turn leads to the formation of a single large outer loop consisting of small loops. The next step is to find all possible variants of crossingings between neighboring loops, that is, the formation of a loop intersection structure.
The third step in this process is the representation of all the structures obtained as a set of nodes, and all crossing-overs as edges of the future structure.
Next, the covering tree of the double graph is calculated, after which it is converted into the ssDNA framework. This process is completely automatic. Finally, a three-dimensional structural model is generated at the atomic level.
The image 2B shows two options for forming the required geometry of the structure: with a discrete edge length or with a continuous edge length. The second option allows you to create a structure with an arbitrary edge length, which in turn makes it possible to get the greatest folding. These variations of the structuring of the geometry were verified by means of a scanning atomic force microscope (images on the right of image 2B ).
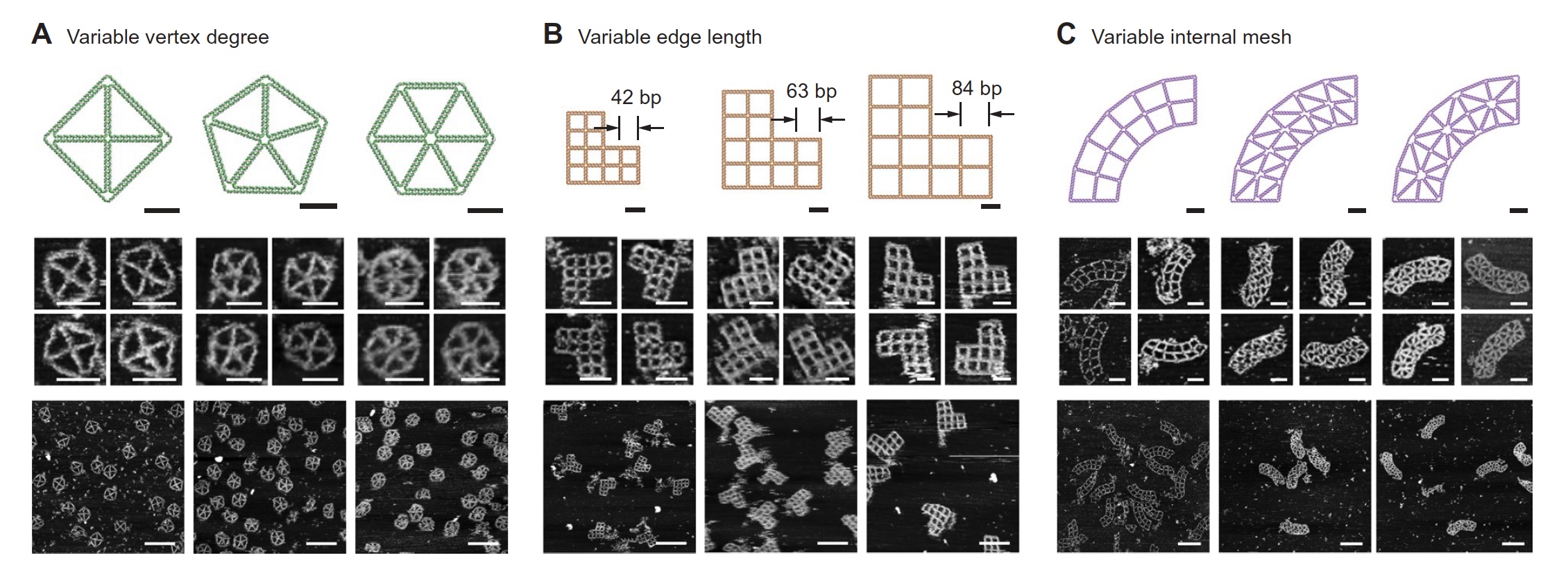
Image number 3
The researchers also set a goal to examine in more detail such aspects of the target geometry as mechanical rigidity, shape accuracy, structure size, internal mesh size and types of its elements. It was also necessary to understand how important the N-branches play, that is, the number of connections between the ribs and, as a result, the overall complexity of the structure. Analysis of the AFM data showed that two-dimensional structures with a large number of branches and a longer edge (due to unpaired nucleotides) are obtained fairly well formed, and the heterogeneity of particles in their structure is minimal.
The internal grid of the structure had three options: triangles with the same direction, squares and triangles with different directions ( 3C ). It was the model with triangles that showed the greatest accuracy in the formation of the required geometry, in contrast to the model with squares. The presence of mixed directions in the triangular model, so to speak, allowed us to obtain more symmetric N-branches and a more accurate form (less distorted).
Scientists note that the analysis of the internal grid of the structure being modeled is extremely important for understanding the mechanical rigidity of this structure. And it is exactly the model described above that is ideal for creating sufficiently strong, accurate (in terms of compliance with the given parameters) and flexible structures.
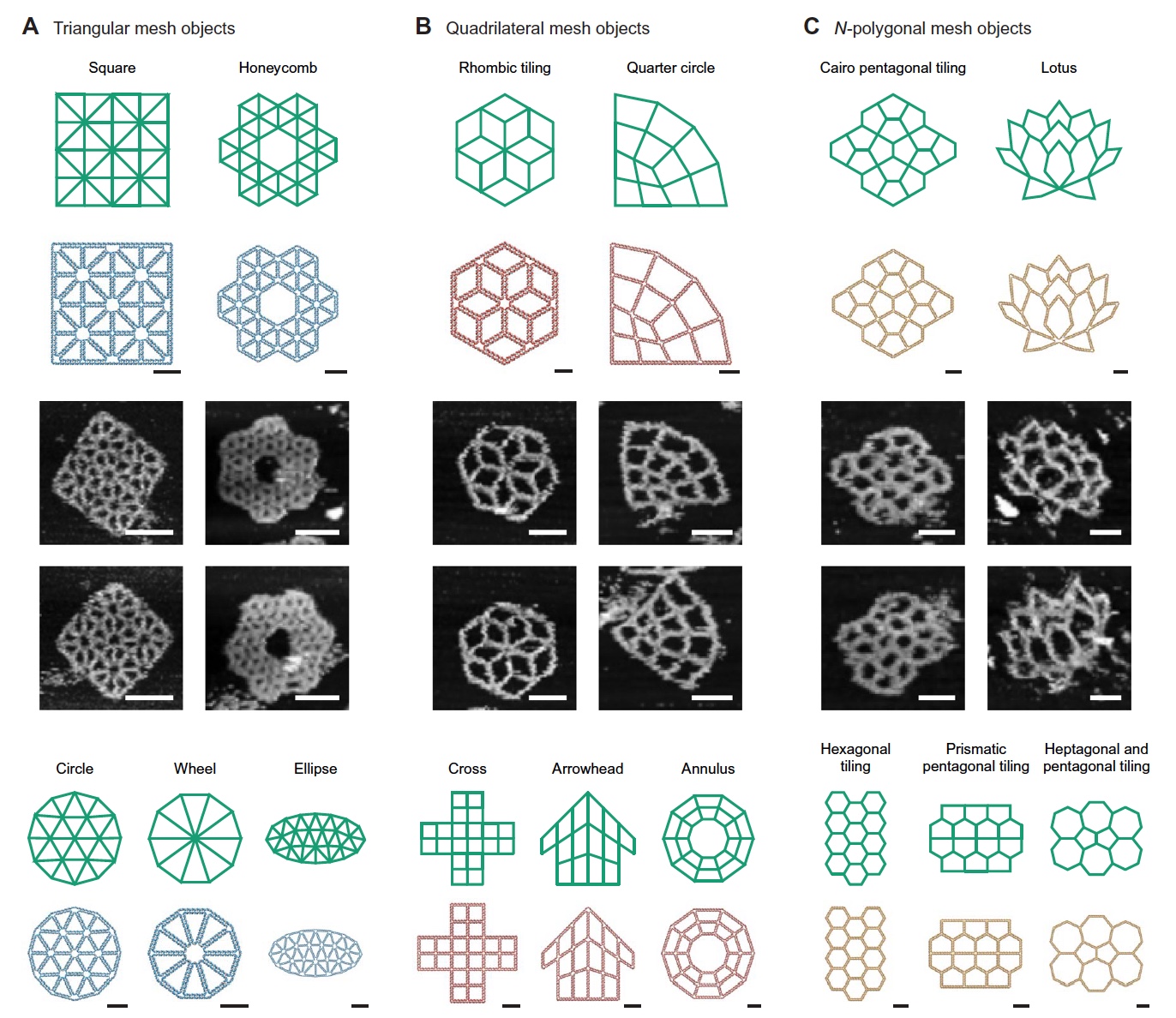
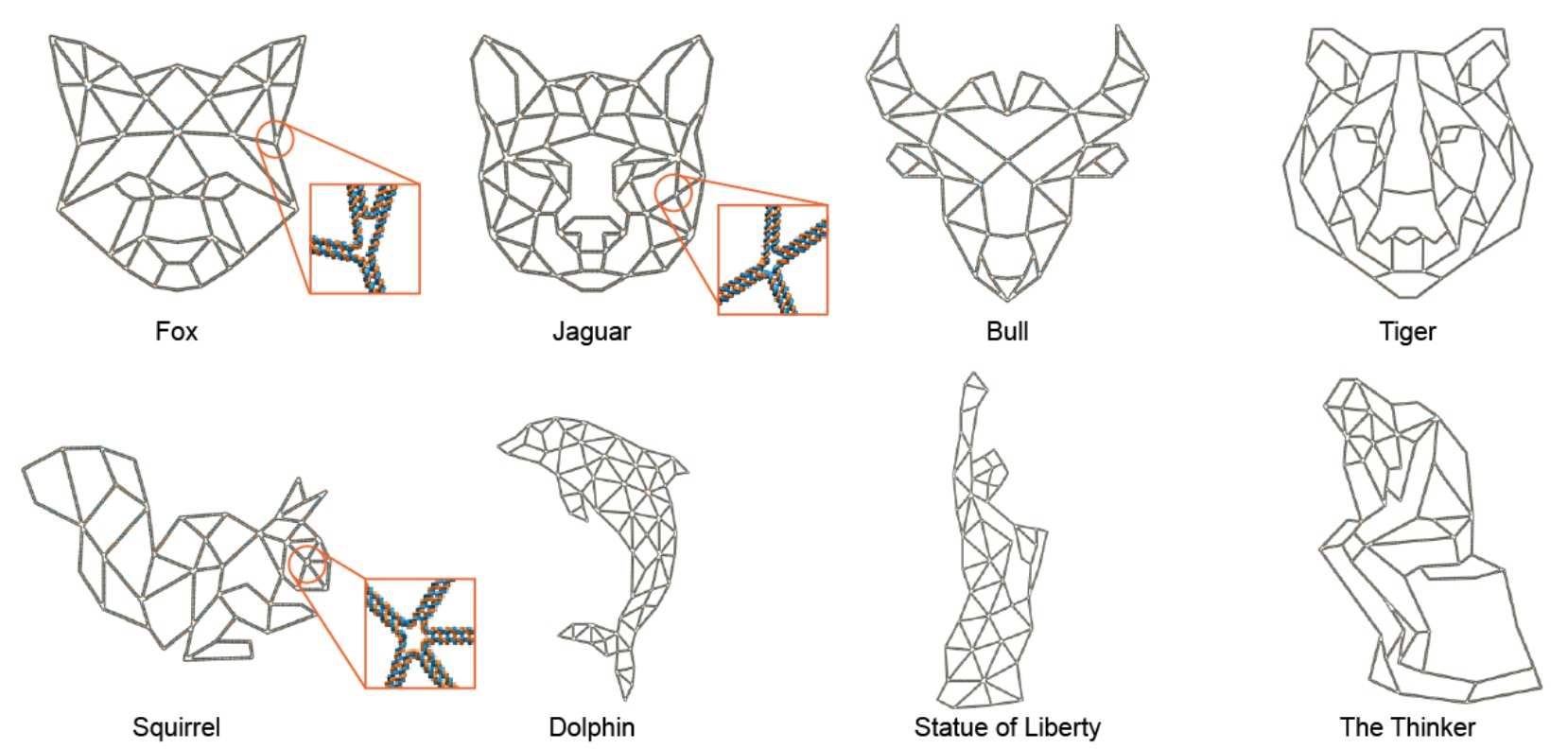
Image number 4
Of course, scientists are not without a sense of beauty, so to speak. Therefore, they decided to demonstrate the possibilities of their development by designing structures of 15 different forms, more precisely with different internal grids and frame. We can see the variants in the image above: here there are “ordinary” squares, a quarter of a circle, and even a lotus.
Open source software
As I mentioned earlier, the work of researchers is open to everyone by reference (), where you can find the necessary program elements (MATLAB, Python 2.7 and Shapely 1.6.4).
As for the algorithm of forming the geometry of DNA origami structures, the instructions for downloading and launching it are presented below in video format. All videos I hide under the spoiler, so as not to stretch the article.
Video # 1: PERDIX launch
The link for downloading the necessary files that was in the video is not working now. Here is an alternative - download link .
The link for downloading the necessary files that was in the video is not working now. Here is an alternative - download link .
Video # 2: Designing the frame (perimeter) of the structure in PERDIX
Video # 3: Designing the frame and internal structure in PERDIX
Video №4: atomic models (different grids)
Video # 5: Atomic Models (N-Branches)
Video # 6: Atomic Models (L-model)
Video №7: atomic models (curved "branch")
For more detailed acquaintance with this study and its results, I strongly recommend to look into the report of the research group and additional materials to it.
Epilogue
This study was aimed at simplifying a fairly time consuming and process-intensive process. And it turned out. The PERDIX algorithm allows you to create structures with the most different geometry, while setting the minimum necessary parameters. DNA has long been the object of study by scientists, not only as part of all life and information carrier, but also as a possible variant of the basis of future technologies. Such works make it possible to understand in more detail (and clearly) how wide the possibilities of nanostructures based on DNA origami are.
The second pleasant moment in this study, at least for me, is that the algorithm is available to everyone. Anyone can use it, anyone can improve it. By giving full access both to their report and to the software itself, scientists not only contribute to the popularization of their research industry, but also to science as such.
I have come across articles in which scholars have spoken quite critically about a paid system of access to reports, arguing that this puts an additional barrier between knowledge and ordinary people. The situation in this matter is very ambiguous and controversial, because everyone should earn money (scientific publications, research groups and scientific sites), but also the strong increase in the cost of access to some research makes them inaccessible to ordinary science lovers, students and even some professors and scientific figures. As I have already said, the question is ambiguous, therefore we will not touch it strongly. In any case, we are pleased that our heroes of today have provided us with full access to their work, which deserves not only attention, but also frank admiration.
Thank you for your attention, remain curious and have a good working week, guys.
Thank you for staying with us. Do you like our articles? Want to see more interesting materials? Support us by placing an order or recommending to friends, 30% discount for Habr users on a unique analogue of the entry-level servers that we invented for you: The whole truth about VPS (KVM) E5-2650 v4 (6 Cores) 10GB DDR4 240GB SSD 1Gbps from $ 20 or how to share the server? (Options are available with RAID1 and RAID10, up to 24 cores and up to 40GB DDR4).
VPS (KVM) E5-2650 v4 (6 Cores) 10GB DDR4 240GB SSD 1Gbps until spring for free if you pay for a period of six months, you can order here .
Dell R730xd 2 times cheaper? Only we have 2 x Intel Dodeca-Core Xeon E5-2650v4 128GB DDR4 6x480GB SSD 1Gbps 100 TV from $ 249 in the Netherlands and the USA! Read about How to build an infrastructure building. class c using servers Dell R730xd E5-2650 v4 worth 9000 euros for a penny?
Source: https://habr.com/ru/post/436116/